Day 50
Aug 16, 2017 · 965 words
Assigning the same data to various objects shouldn’t pose a memory issue, as in Python variables store references to data so only one instance of the source data is actually stored. You can verify this with the built-in id() method to see where variables point to. There are still some areas I’ve have to optimize in the multi-column model though.
As I pointed out yesterday, I don’t need to run the window calculations for every column. Even if two objects run the same calculations on a variable, they will point to different areas of memory (so in that case you would be storing duplicates of the same object). So once I set those attributes for the forests, I want to make sure the forests don’t try to do any further manipulation to those variables.
Reorganizing the classes
Since the classes for single and multi random forest models will share a lot of attributes / methods, it would make more sense to start with a BaseModel class from which the SingleRFModel and MultiRFModel classes branch off of.
Right now, BaseModel holds a majority of the attributes and methods. SingleRFModel and MultiRFModel just have their own versions of the train() and predict() methods since that’s the only area where they really differ. SingleRFModel.predict() returns the values and standard deviations as Series, while MultiRFModel.predict() returns them as DataFrames (with columns labeled accordingly). That means the structure of the predictions should mirror the structure of the original data.
Here is the current code for MultiRFModel:
class MultiRFModel(BaseModel):
def __init__(self, *args, **kwargs):
super().__init__(*args,**kwargs)
self.columns = list(map(lambda col: self.data[col], self.data))
self.estimators = list(map(lambda col:
RandomForestRegressor(n_estimators=self.n_estimators),
self.data))
def train(self):
for i,rf in enumerate(self.estimators):
rf.fit(*self._get_training_arrays(self.columns[i]))
def predict(self):
pred_start_date = self._get_pred_start_date(self.data)
preds = list(
map(lambda rf,s: self._get_prediction(rf,s,pred_start_date),
self.estimators,self.columns))
arr_vals, arr_std = list(zip(*preds))
return (pd.concat(arr_vals,axis=1),
pd.concat(arr_std,axis=1))
You can find the full code including BaseModel in the energize module.
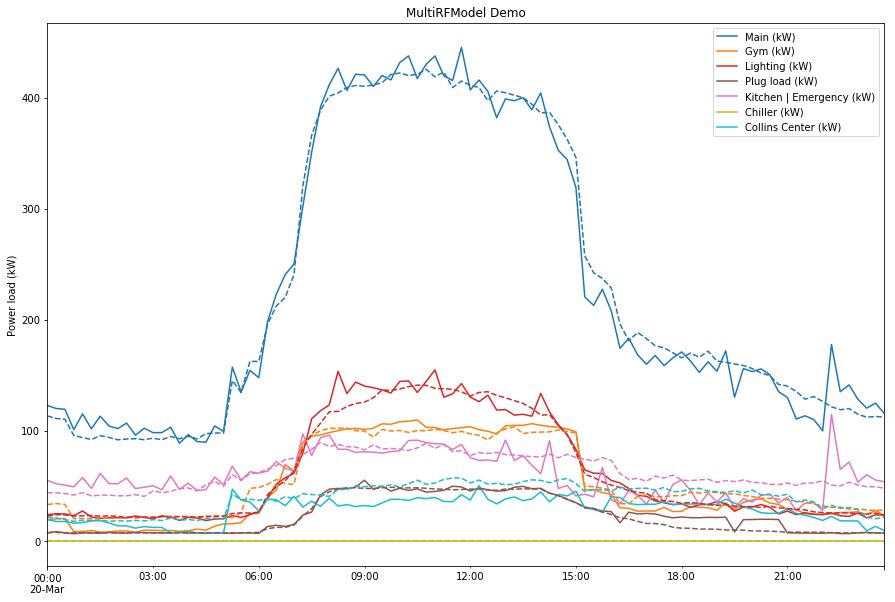
Here’s an example of using a MultiRFModel to run the same type of prediction that I’ve done in the past, but on all of the circuit panels in df_energy. The solid lines are the actual values (not fed into the model) and the dotted lines are the predictions.
Not pictured are the thresholds, but the data for them is calculated. The entire process takes about 10 seconds to run. That includes all of the following:
- Reading the source data
- Creating the training windows
- Building the training arrays
- Auto-upsampling the extra features
- Training all 7 of the forests (each forest contains 100 decision trees)
- Calculating the expected values and standard deviations for each column
Pretty amazing, especially considering the old hardware I’m running on.
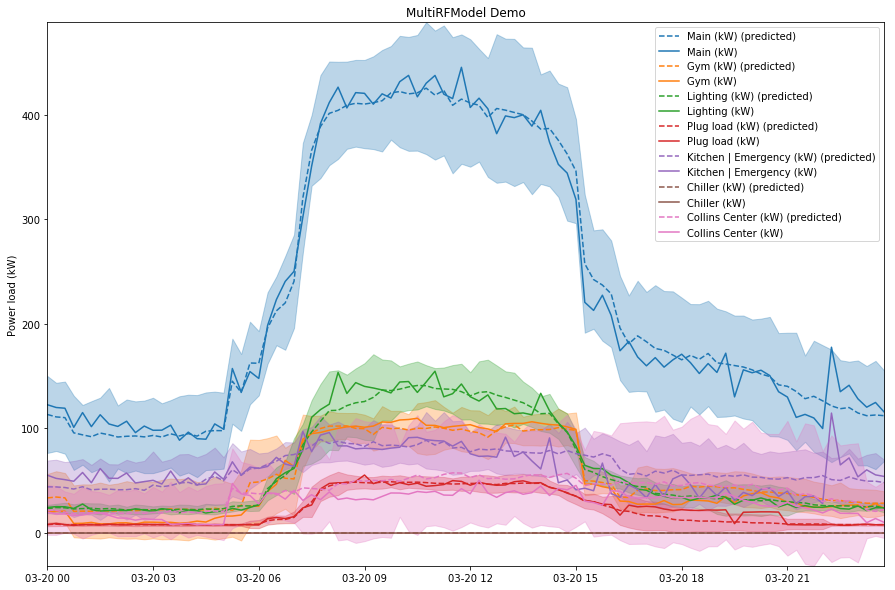
Here’s another plot that shows the predictions above but with $2\sigma$ threshold regions added. It’s pretty cluttered but you get the idea:
In practice you would care mostly about the top of the threshold, and you would probably only look at one panel at a time rather than all of them overlayed.
I’m really pleased with the status of the model right now. It’s incredibly straightforward to now set up, whereas previously you had to use a much longer script to set up all the features manually and tinker with things until it worked.
A quick note on temperature features
I experimented with adding the temperature data as a feature and found that it did not improve the predictions (from my anecdotal test it provided a worse fit than without including it).
Appending to a CSV file
I know by default the to_csv() method will just write the data contents to the file at the given path, overwriting it. I want my script to add the new predictions to the end of the prediction table. This post explains how to do that by setting the mode argument to 'a' for append.
I also wonder if I should have a third table that contains metadata about the predictions (i.e. the model attributes), or at least some kind of text log. That way if someone decides to change the parameters of the prediction process, then the previous prediction will have some context as to how they were made (so you don’t mistakenly view all predictions as equal). For example, it could contain the input/gap/output sizes, sample frequecy, number of estimators, extra feature names, and time attributes to name a few.
I could compromise and make a log stored in a CSV file, so each time it predicts it adds a single entry to the log with each column holding a different meta-attribute.
Running a trial
To really put this to the test I’m going to make a new data file, this time with the final week of power data removed. I’ll then make two more files, each with one more day of data than the last. For each file I’ll make a model and add its predictions to a continuous prediction table. Then I’ll pull in the prediction table and verify that it lines up with the actual values.
I set up the headers for the value and variance prediction tables as follows:
pd.DataFrame(columns=df_data.columns).to_csv('resources/test_files/vals.csv')
pd.DataFrame(columns=df_data.columns).to_csv('resources/test_files/std.csv')
The predictions were made with:
def make_pred(num):
base_path = 'resources/test_files/'
df = pd.read_csv(base_path+'file'+str(num)+'.csv',index_col=0)
df.index = pd.to_datetime(df.index)
df = egz.only_full_days(df,'15min')
df.index.freq = egz.inferred_freq(df)
model = egz.MultiRFModel(
data = df,
td_input = pd.Timedelta(weeks=4),
td_gap = pd.Timedelta(days=1),
td_output = pd.Timedelta(days=1),
time_attrs = ['dayofyear','dayofweek'],
extra_features=pd.DataFrame(df_extras['noschool']),
n_estimators=50)
model.train()
vals, std = model.predict()
vals.to_csv(base_path+'vals.csv', mode='a', header=False)
std.to_csv(base_path+'std.csv', mode='a', header=False)
Then I just ran a loop calling the above function on all 4 test files I made.
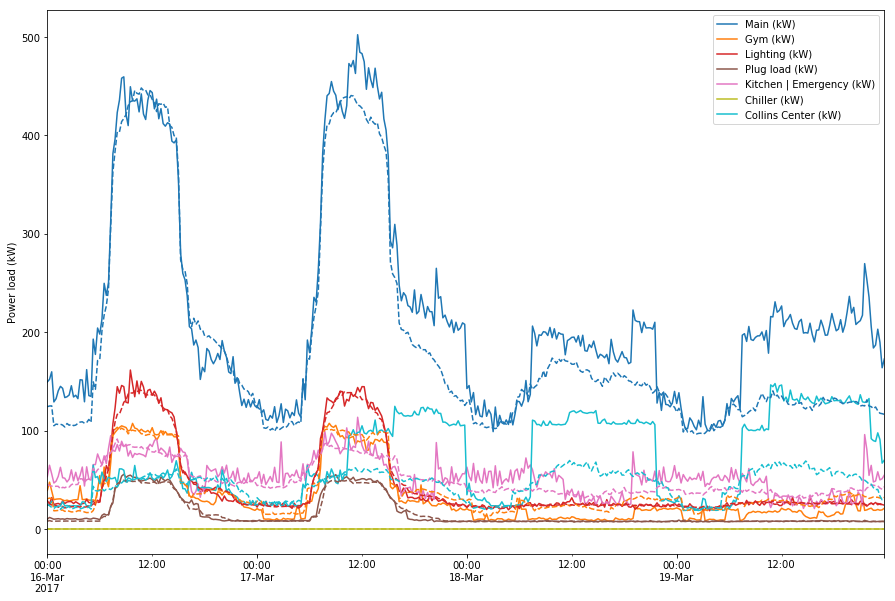
Semi-success
I was able to append the predictions to the value and variance tables.
This set of predictions looks off compared to the predictions I’ve seen so far. When I use my old script to predict the 19th for example, I get a much closer fit that what is pictured here.
I’ll have to figure out what the problem is tomorrow.