Day 49
Aug 15, 2017 · 807 words
Getting the variance
For clarity’s sake I think I’ll keep the variance output seperate from the prediction output. They should be stored in two seperate tables, one for predictions and one for variance. However instead of making two seperate functions (which would have some overlap of calculactions) I can run the calculations in the same function and return a tuple of the two Series.
I’ll use the same technique from before, getting the standard deviation of the residuals of the forest’s decision trees:
def _get_pred_std(self,X):
all_pred = np.array([t.predict(X) for t in self.rf])
return np.std(all_pred,0).ravel()
This is called from the _get_prediction method of the class (called from the public predict method) which then returns a tuple of the predicted power values and the predictions’ standard deviations.
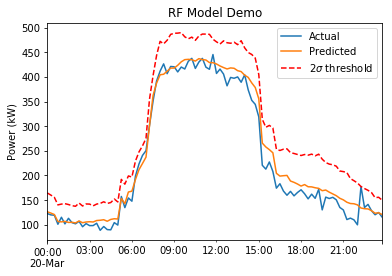
model.train()
vals,std = model.predict()
data[vals.index].plot(title='RF Model Demo', label='Actual')
vals.plot(label='Predicted')
(vals + std*2).plot(label='$2\sigma$ threshold', style='r--')
plt.ylabel('Power (kW)')
plt.legend()
That demonstrates the updated process of training the model and using predictions to make a sample plot:
Issue: changing window sizes
I found that changing the output window size throws some errors in my tests. The main issue was in the pred_start_date calculation which I resolved. Another issue was that it was possible to get null values when calling resample().asfreq() but this did not occur when I replaced it with just asfreq(). Now I can generate larger output periods. The reason I was getting errors on smaller windows sizes was because my extra_features table was not appropriately upscaled.
I think I found a convenient way to automatically upscale the extras features if necessary:
extras.fillna(extras.asfreq(timedelta(self.td_output)).ffill())
That only upsamples rows that have missing values where values are needed. I moved this process into a _validated_feat() method that runs when the RandomForestModel object is initialized.
Now I can safely change all the window parameters regardless of the setup of the feature table and have the predictions run properly.
Reusing models
The current setup implies that the model will be trained and make a single prediction every day. No model is expected to be used more than once. The benefit of this approach is that the model will always be trained on the most up to date information. At smaller output window sizes this requires more computing power as the model has to be trained many times per day, but that’s not the route I’m planning on taking anyways.
Sample weights
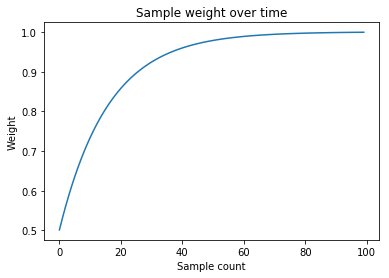
I remember at least one of the research papers I read focusing on giving a higher preference to more recent samples when making a model. At the moment, all samples have the same weight. I want to incorporate some form of weight decay for older samples. I don’t want the dropoff to be too sharp. I think some sort of logarithmic shape is appropriate for the decay. The last sample probably shouldnt’ have zero weight, rather I should set a baseline weight such as 50%. Here’s how I’d generate that region:
1-np.logspace(np.log10(0.5),-3,n_samples)+1e-3)
I’ll probably tweak the baseline weight in the future (I don’t want to discredit too much of the historical data). The extra 1e-3 on the end ensures that the most recent sample has 100% weight. I save the array as sample_weights_. This is a plot of what the region looks like:
Note that here, “sample” means an input vector which contains a window of input data. So the temporal scale of the x-axis depends on your output window size.
Scheduling tasks
To schedule tasks on Linux you can use the built-in cron tool. This post explains how to set up the /etc/crontab file with your desired commands. Before I can set this up, I need to make the actual script that it will run. This moves onto the next order of business:
I drafted up a script that basically does what I’ve already demoed so far except it runs the entire CSV reading process (so each time it runs it accesses the most up to date version of the source data). From there it’s really simple to export a CSV of the predictions with the pandas.Series.to_csv() method.
Todo
At the moment only one circuit panel is considered (I’ve been using the main panel). It would probably be helpful to have predictions for all of the sub-panels as well. To handle this I will most likely want to make another class that works with dataframes and implicity creates a bunch of RandomForestModels for each column (and can return CSV files accordingly). The challenge is to see if I can make a more efficient way to do this. By simply creating a bunch of independent RandomForestModel objects I’m causing a lot of computational overlap. If all the columns occupy the same indicies then the training window index calculation is unecessarily repeated. There is also some overlap of attributes (although I’m pretty sure in Python everything is just stored as pointers anyways so this shouldn’t be a big deal).